**Significant in comparison with 1 nucleolar cells t = 4,27 p < 0,001, with 2 nucleolar cells
·Significant in comparison with 1 nucleolar cells t = 2,64 p < 0,05
··Significant in comparison with 1 nucleolar cells t = 2,80 p < 0,01
The difference between minimal and
maximal meanings of the DNA quantity as in the nucleus as in nucleolus, at
control and experiment was insignificant. Table 2
Under action of a virus there are no
significant changes in the DNA quantity in a nucleus of HEp-2 cells (180,8±28,1 in the control, 178,8±31,4 at
a virus infection). The area of nucleuses also remains without changes
(64,6±9,8 - control and 64,9±10,2 - OPV infection). The difference in (among) DNA
quantity into "total" nucleolus also is absent (27,46±3,4 - control, 26,26±3,2 -
infection).
As it follows from the tables 1 and 2 the
difference of the DNA quantity between separate populations of HEp-2 cells was
often noticeably (the comparison of 4 and 2 nucleolar cells t=1,99 p < 0,05).
So in cells of one line, in nucleus with different numbers of nucleoli we
defined high difference in DNA quantities. It is probably can be explained by
the genotipically realization of the NORs number.
|
Number of ucleolus in the nucleus |
% of the cells in population* |
Nucleus |
Summarized nucleolus |
||||
|
quantity of DNA (in C.U.) |
area |
perimetr |
quantity of DNA (in C.U.) |
area |
perimetr |
||
|
1 |
12,7 |
155±25 |
60±12 |
18±3,4 |
20,2±2,7 |
7,4±0,8 |
4,9±1,1 |
|
2 |
35,5 |
178±30 |
65±12 |
19±3 |
23,4±4,8 |
8,5±1,7 |
7,1±1,1 |
|
3 |
25,1 |
168±23 |
61±7,7 |
18±2,3 |
27,4±7,9 |
10±2,9 |
8,9±2,1 |
|
4 |
22,5 |
197±24 |
70±9,5 |
18,6±5 |
31,0±3,8 · |
12±1,5 |
13,1±1,1 |
|
5 and more |
4,2 |
224±25 |
81±11 |
20±5,1 |
36,1±4 · · |
13±2,6 |
14,0±1,7** |
*Without
the account the mitosis, dead and nonnucleolar cells
**Significant
in comparison with 1 nucleolar cells t = 4,48 p < 0,001 with 2 nucleolar cells
t = 3,39 p < 0,01 with 3 nucleolar cells t = 2,15 p < 0,05
fSignificant in comparison with 1 nucleolar cells
t = 5,26 p < 0,001 with 2 nucleolar cells
t = 3,85 p < 0,01
·Significant in comparison with 1 nucleolar cells
t = 2,31 p < 0,05
··Significant in comparison with 1 nucleolar cells
t = 4,99 p < 0,001 with 2 nucleolar cells
t = 1,98 p = 0,05
Our experiments showed that with the
increasing of the nucleoli quantity the quantity of the DNA in individual
nucleolus decreases, but the quantity of the total DNA total nucleoli has the
tendency to increase in HEp-2 cells both in the control, and under the action of
OPV.
From our data increasing of the nucleolus
number in nucleus the DNA quantity in individual nucleoli decreases, but the
quantity of DNA in "total" nucleolus has the tendency to increase in HEp-2
resowing cell line both in the control, and under the action OPV.
According to [27] informative parameter of
the cells differentiation is the area of a nucleus. From our experiments were
received the same area of a nucleus both in control 65,6±7,8, and during infection of OPV 64,9±8,1. Also there were not the changes of the nuclear
perimeter (16,8±2,8 - control 18,3±3,3 - infection).
We investigate possible changes in nucleolar
indices. They are almost the same - nucleolus area (9,85±1,1 in control, 9,79±1,8 - virus
action) and perimeter 7,57±1,6 - control 8,91±2,1 - action of OPV).
Also we investigated the concentration of the
DNA in a nucleus. This index was characterized by constancy with very few
deviations in control (2,79±0,02). The concentration of
the DNA wasn't changed under the influence of OPV but the deviation was
increased (2,75±0,11). Also the DNA concentration in
the nucleolus wasn't changed (2,79±0,04 - intact cells,
2,69±0,08 experience).
|
Number of nucleolus in the nucleus |
Nucleolus/nucleus |
||
|
DNA |
area |
Perimeter |
|
|
1 |
0,13±0,03 |
0,13±0,02 |
0,31±0,05 |
|
2 |
0,17±0,02 |
0,17±0,03 |
0,45±0,04 |
|
3 |
0,16±0,03 |
0,16±0,03 |
0,48±0,07 |
|
4 |
0,16±0,04 |
0,16±0,04 |
0,63±0,09* |
|
5 and more |
0,15±0,03 |
0,15±0,03 |
0,64±0,04* |
*Significant in
comparison with 1 and 2 nucleolar cells t = 5,15, t = 3,3, p < 0,001, in
comparison with 3 nucleolar cells t = 1,98, p <
0,05
**Significant in comparison with 1 nucleolar cells t = 3,1 p
< 0,01
|
Number of nucleolus in the nucleus |
Nucleolus/nucleus |
||
|
DNA |
area |
Perimeter |
|
|
1 |
0,13±0,03 |
0,13±0,03 |
0,27±0,07 |
|
2 |
0,13±0,02 |
0,13±0,02 |
0,39±0,07 |
|
3 |
0,16±0,04 |
0,17±0,04 |
0,5±0,1 |
|
4 |
0,16±0,03 |
0,17±0,03 |
0,73±0,1* |
|
5 and more |
0,16±0,04 |
0,16±0,02 |
0,71±0,1** |
*Significant in
comparison with 1 nucleolar cells t = 3,6 p < 0,001, with 2 nucleolar t = 2,6, p
< 0,01
**Significant in comparison with 1 nucleolar cells
t = 3,77, p < 0,001, with 2 nucleolar t = 2,79, p < 0,01
From tables 3 and 4 we see that the DNA
quantities in the nucleolus/nucleus ratio do not depend from the nucleolus
number in the nucleus in the control (average meaning in population 0,156±0,014) and in infection condition (average meaning in
population 0,151±0,021). These data were received with
taking account of the percent of each type of cells in the population (tables 1
and 2). The significant difference in the nucleolus/nucleus ratio was absent not
only in population, but also in individual cells.
According to the data of tables 3 and 4 the
essential difference in HEp-2 cells with various quantity of nucleolus in
nucleus is the sums of perimeters of the "total" nucleolus in the nucleus. These
data demonstrate almost direct linear dependence on the increasing of the
nucleolus quantity both in experience and in the control.
Our data demonstrate the absence of changes
of the total nucleolar DNA with the increasing of the number of nucleolus in the
nucleus while quantity individual nucleolar DNA was appreciably decreased in
process of the increasing of nucleolus in the nucleus.
|
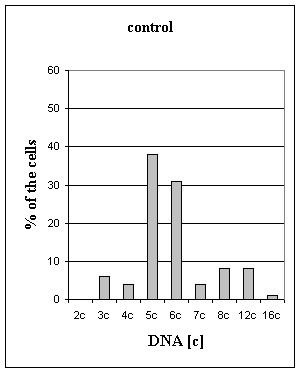
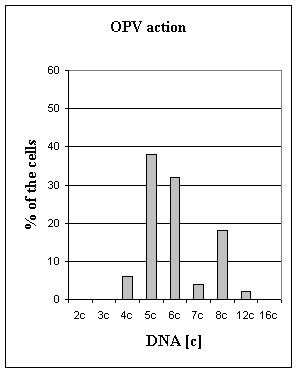
Figure 1. Distribution of the nucleus by the DNA ploidy (in "c" units)
in HEp-2 cells
Fig 1 summarizes changes in DNA ploidy
indices in normal condition and under the OPV action. The present results
indicate that under the action of OPV significant changes in ploidy of HEp-2
cells were absent (6,03 "c" in control 5,96 "c" - OPV infection). Percentage of
aneuploid cells was decreased, and percentage of euploid cells was increase
(13±4,1 in control 24±3,2 OPV
action, t=2,11, p < 0,1). Institute of molecular biology NSA RA
We have shown that in HEp-2 cell line
quantity of the nucleolus does not depend on the quantity of the DNA in a
nucleus, both at action of a virus, and in intact cells HEp-2. Quantity of the
DNA in the nucleolus in direct proportion with the quantity of the DNA in a
nucleus, both in experience, and in the intact HEp-2 cells. Conducting the
nucleoli cytometry in the intact cells and under influence of the OPV, the
relation of the sums of the nucleolar perimeters in a nucleus is the significant
factor. It increases/decreases linearly while number of nucleoluses
increases/decreases in a nucleus. Was shown the increase of the summarized
nucleolar DNA at the increase of the nucleolus number in a nucleus of HEp-2
cells more expressed at action of a virus. Also was shown the reduction of the
DNA quantity individual nucleoluses in process of growth of the number of
nucleolus in HEp-2 cells.
There was tendency to the increasing of the
cells number with euploid DNA quantity in infected cell line in comparison with
control.
The infected cell line had the tendency
increases the number of cells with euploid quantity of the DNA in nucleus in
comparison with control. These data allow to assume, that under the influence of
OPV there was the a decrease of a proliferation activity of HEp-2 cells.
1. Field D., Fitzerald P., Sin F.
- Cytobios. 1984. V. 41. P.
23-33.
2. Smetana K., Bush H.
- In: The cell nucleus. N. Y. 1975. P.
126-144.
3. Derenzini M., Trere
D., Pession A., Montanaro L., Sirri V., Ochs R. L. -
American Journal of Pathology. 1998. V. 152. P.
1291-1297.
4. Ceccarelli C.,
Trere D., Santini D., Taffurelli M., Chieco P, Derenzini M., K, Dodge R., Barsky
S.H. - Arch Pathol Lab Med.2000. 124(2).
P.221-7.
5. Hernandez-Verdun D.,
Derenzini M. - Eur. J. Cell Biol. 1983. V. 31. P.
360-365.
6. Troster H., Spring
H., Meissner B., Shultze P., Trendenburg M. F. - Chromosoma.
1985. V. 91 P. 151-163.
7. Derenzini M., Pession A., Farabegoli F., Trere D., Badiali M., and Dehan
P. - American Journal of Pathology. 1989. V. 134. P.
925-932.
8. Mirre C., Knibiehler
B. - J. Cell Sci. R. 1982. V. 55. P.
261-276.
9. Mirre C., Knibiehler
B. - Protoplasma. 1984. V. 121. P.
120-128.
10. Howell W.
M. - Chromosoma. 1977. V. 62. P.
361-7
11. Canet V, Montmasson MP,
Usson Y, Giroud F, Brugal G. - Cytometry. 2001. V. 43(2). P.
110-6.
12. Derenzini M.,
Farabegoli F., and Trere D. - J. Histochem. Cytochem. 1993.
V. 41. Issue 6. P. 829-836.
13. Derenzini M., Trere D., Pession A., Govoni M., Sirri V., Chieco P.
- J. Patholl. 2000. V. 191(2). P.
181-186.
14. Roussel P., Andre
C., Comai L., Hernandez-Verdun D. - J. Cell Biol. 1996. V.
133. P. 235-246.
15. Baak J.P.
- Pathol. Res. Pract. 1984. V. 179(2). P.
193-199.
16. Friedrich K.,
Scheithauer J., Dimmer V., Meyer W., Theissig F., Haroske G., Kunze K. D.
- Anal. Cell. Pathol. 2000. V20(2-3). P. 69-82.
17. Silvestrini R. - Annals of Oncology. 2000. V. 11. P.
259-261.
18. Kado G. - Dev. Biol. Stand. 1976. V. 37. P.
261-264.
19. Böcking A, Adler CP, Common HH, Hilgarth HM, Granzen B, Auffermann
W. - Analyt. Quant. Cytol. 1984. V. 6. P. 1-8.